Motifs detected by DREME on the MeRIP-seq datasets
4.6 (779) · € 17.50 · En Stock
Download scientific diagram | Motifs detected by DREME on the MeRIP-seq datasets from publication: A novel algorithm for calling mRNA m 6 A peaks by modeling biological variances in MeRIP-seq data | Motivation: N⁶-methyl-adenosine (m⁶A) is the most prevalent mRNA methylation but precise prediction of its mRNA location is important for understanding its function. A recent sequencing technology, known as Methylated RNA Immunoprecipitation Sequencing technology (MeRIP-seq), | mRNA, RNA Sequence Analysis and Base Sequence | ResearchGate, the professional network for scientists.

CUCUME: An RNA methylation database integrating systemic mRNAs signals, GWAS and QTL genetic regulation and epigenetics in different tissues of Cucurbitaceae - ScienceDirect

MeTDiff: A Novel Differential RNA Methylation Analysis for MeRIP-Seq Data

Motifs detected by DREME on the MeRIP-seq datasets

Motifs detected by DREME on the MeRIP-seq datasets

Comprehensive analysis of transcriptomic profiling of 5‐methylcytosin modification in placentas from preeclampsia and normotensive pregnancies - Wei - 2023 - The FASEB Journal - Wiley Online Library

Comprehensive analysis of N6-methyladenosine-related RNA methylation in the mouse hippocampus after acquired hearing loss, BMC Genomics
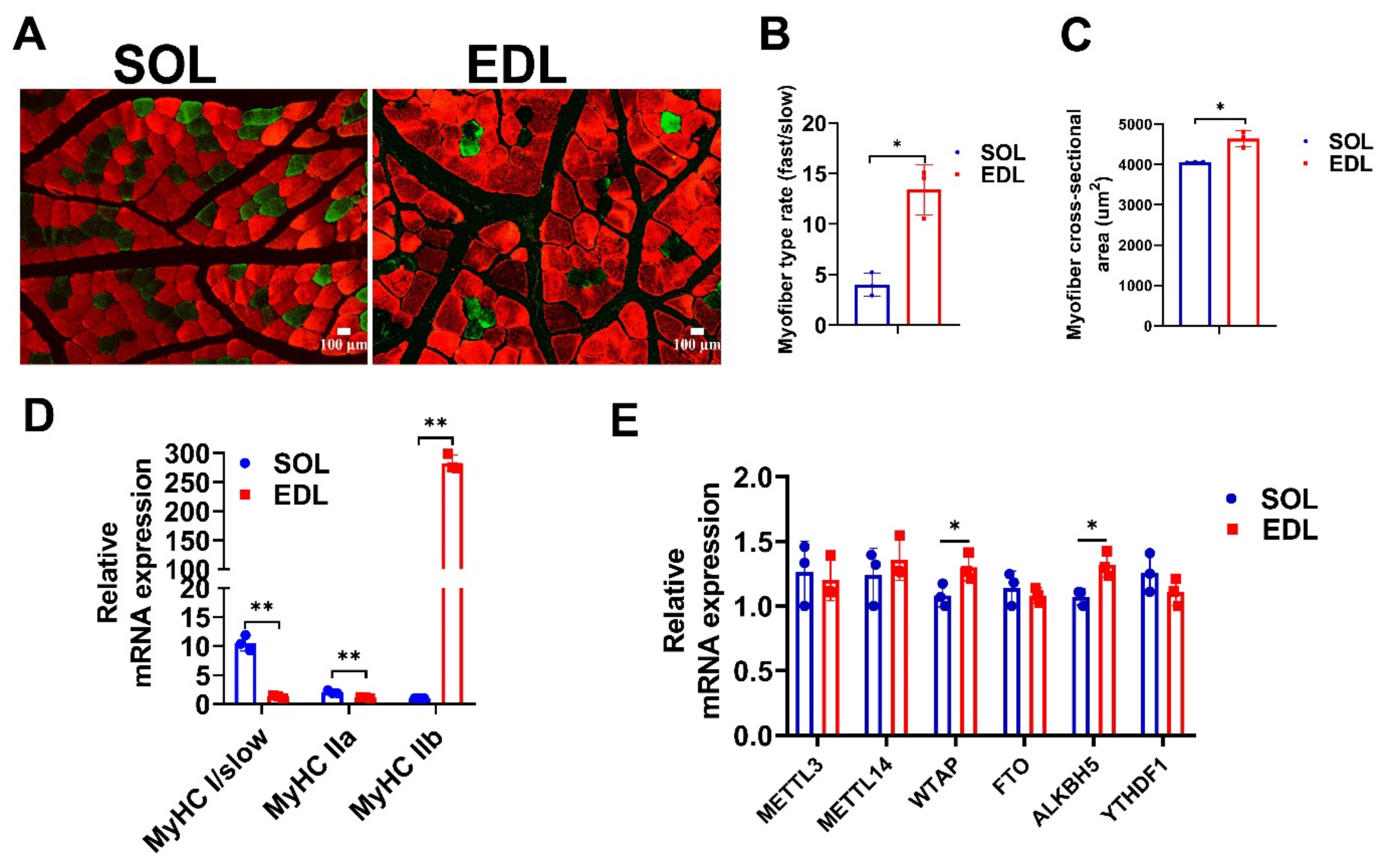
IJMS, Free Full-Text

The Role of m6A/m-RNA Methylation in Stress Response Regulation - ScienceDirect

A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package - ScienceDirect











